
Students
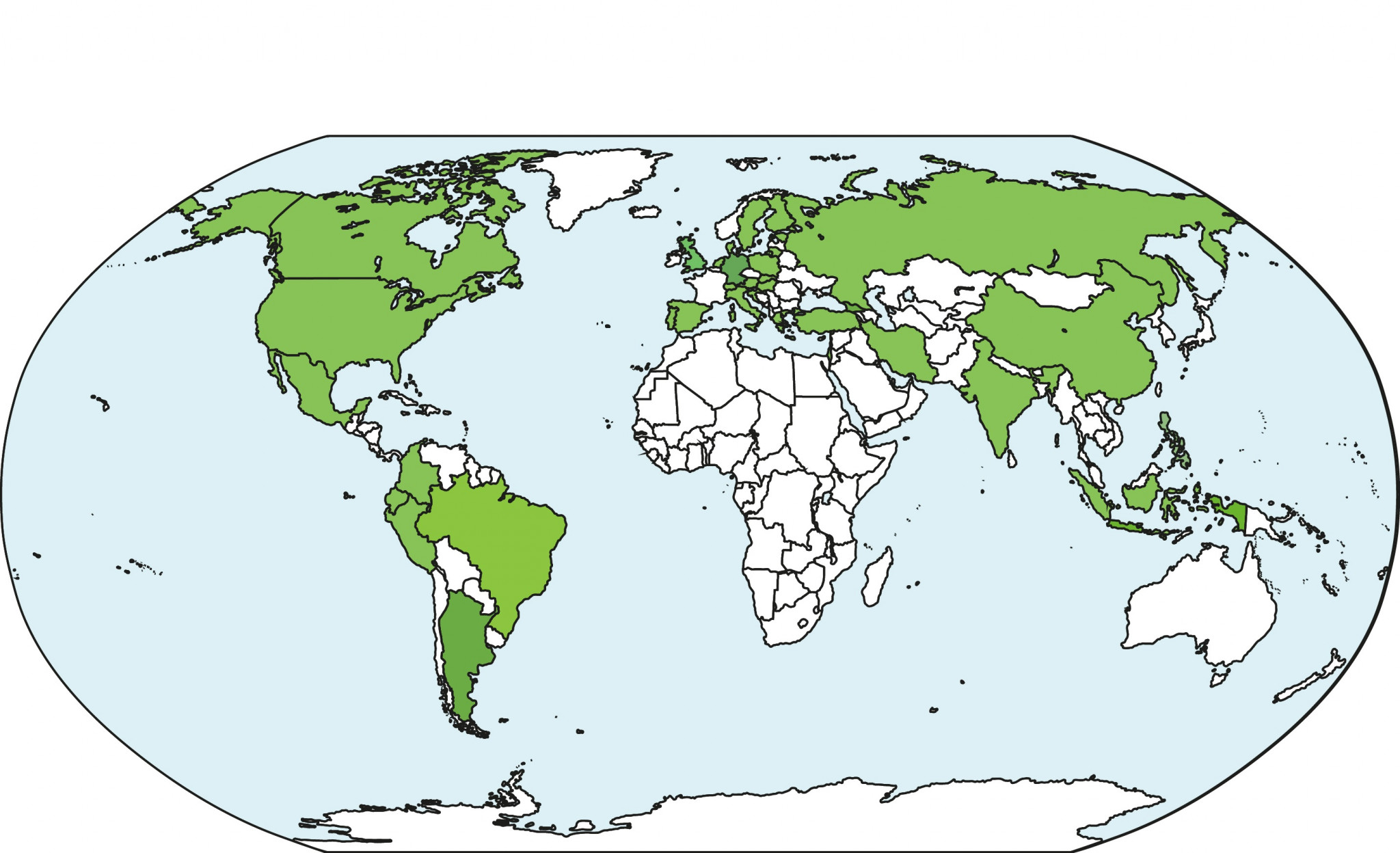
As of January 2026, QMB has 45 enrolled graduate students and approximately 100 alumni. Half of our students have an international background, representing 15 different nationalities, and gender representation is evenly balanced.
 Wasim Aftab (Alumnus)
Wasim Aftab (Alumnus)

Wasim Aftab (Alumnus)
| Class of 2015
Country of origin: India
MSc: Computer Engineering, King Abdulaziz University Jeddah, Saudi Arabia
PI: Axel Imhof, Medical Faculty, LMU Munich
CoPI: Ralf Zimmer, Faculty of Mathematics, Informatics and Statistics, LMU Munich
Project: Computational methods for exploratory analysis of proteomics data
2022
W Aftab, S Lahiri, A Imhof: An open-source software for probabilistic identification of proteins in situ and visualization of proteomics data, MCPRO 100242
View publication2021
A. Lukacs, A. W. Thomae, P. Krueger, T. Schauer, A. V. Venkatasubramani, N. Y. Kochanova, W. Aftab, R. Choudhury, I. Forne & A. Imhof: The Integrity of the HMR complex is necessary for centromeric binding and reproductive isolation in Drosophila, https://doi.org/10.1371/journal.pgen.1009744
View publicationLahiri S, Aftab W, Walenta L, Strauss L, Poutanen M, Mayerhofer A, Imhof A.; Life Sci Alliance. 2021 Jan 6;4(3):e202000672. doi: 10.26508/lsa.202000672. Print 2021 Mar. PMID: 33408244; MALDI-IMS combined with shotgun proteomics identify and localize new factors in male infertility.
View publicationLahiri S, Aftab W, Walenta L, Strauss L, Poutanen M, Mayerhofer A, Imhof A. Life Sci Alliance. 2021 Jan 21;4(3):e202101015. doi: 10.26508/lsa.202101015. Print 2021 Mar. PMID: 33479050; Correction: MALDI-IMS combined with shotgun proteomics identify and localize new factors in male infertility.
View publicationAftab W, Imhof A.; Adv Exp Med Biol. 2021;1336:105-128. doi: 10.1007/978-3-030-77252-9_6. PMID: 34628629; Discovery of Native Protein Complexes by Liquid Chromatography Followed by Quantitative Mass Spectrometry
View publicationHayn M, Hirschenberger M, Koepke L, Nchioua R, Straub JH, Klute S, Hunszinger V, Zech F, Prelli Bozzo C, Aftab W, Christensen MH, Conzelmann C, Müller JA, Srinivasachar Badarinarayan S, Stürzel CM, Forne I, Stenger S, Conzelmann KK, Münch J, Schmidt FI, Sauter D, Imhof A, Kirchhoff F, Sparrer KMJ.; Cell Rep. 2021 May 18;35(7):109126. doi: 10.1016/j.celrep.2021.109126. Epub 2021 Apr 27. PMID: 33974846; Systematic functional analysis of SARS-CoV-2 proteins uncovers viral innate immune antagonists and remaining vulnerabilities
View publication2020
Singh AP, Salvatori R, Aftab W, Aufschnaiter A, Carlström A, Forne I, Imhof A, Ott M.; Mol Cell. 2020 Sep 17;79(6):1051-1065.e10. doi: 10.1016/j.molcel.2020.07.024. Epub 2020 Sep 1. PMID: 32877643; Molecular Connectivity of Mitochondrial Gene Expression and OXPHOS Biogenesis
View publicationSalvatori R, Kehrein K, Singh AP, Aftab W, Möller-Hergt BV, Forne I, Imhof A, Ott; M. Mol Cell. 2020 Feb 20;77(4):887-900.e5. doi: 10.1016/j.molcel.2019.11.019. Epub 2019 Dec 26. PMID: 31883951; Molecular Wiring of a Mitochondrial Translational Feedback Loop
View publicationSalvatori R, Aftab W, Forne I, Imhof A, Ott M, Singh AP. STAR Protoc. 2020 Dec 15;1(3):100219. doi: 10.1016/j.xpro.2020.100219. eCollection 2020 Dec 18. PMID: 33377112; Mapping protein networks in yeast mitochondria using proximity-dependent biotin identification coupled to proteomics
View publicationBasch M, Wagner M, Rolland S, Carbonell A, Zeng R, Khosravi S, Schmidt A, Aftab W, Imhof A, Wagener J, Conradt B, Wagener N.; Mol Biol Cell. 2020 Apr 1;31(8):753-767. doi: 10.1091/mbc.E19-06-0329. Epub 2020 Feb 12. PMID: 32049577; Msp1 cooperates with the proteasome for extraction of arrested mitochondrial import intermediates
View publication Shyam Alagarsamy (until 2020)
Shyam Alagarsamy (until 2020)
Shyam Alagarsamy (until 2020)
| Class of 2020
Country of origin: India
MSc: Molecular Medicine, Univeristy of Goettingen, Germany
Lucas Jae
 Severin Angerpointner
Severin Angerpointner

Severin Angerpointner
| Class of 2021
Country of origin: Germany
MSc: Theoretical and Mathematical Physics, LMU & TU Munich, Germany
PI: Erwin Frey, Statistical and Biological Physics, Dept. of Physics, LMU
2022
A V Dass, S Wunnava, et al., F Gartner, S Angerpointner, E Frey, D Braun: RNA Oligomerisation without Added Catalyst from 2 ′ ,3 ′ ‑Cyclic Nucleotides by Drying at Air‑Water Interfaces; ChemSystemsChem e202200026 (2022)
View publication Žiga Avsec (Alumnus)
Žiga Avsec (Alumnus)

Žiga Avsec (Alumnus)
| Class of 2015
Country of origin: Slovenia
MSc: Biophysics, LMU Munich, Germany
PI: Julien Gagneur, Faculty of Informatics, TU Munich
Project: Systems genetics approaches for rare genetic disorders.
2019
G. Eraslan, Ž. Avsec, J. Gagneur, F. J. Theis, Deep learning: new computational modelling techniques for genomics, Nature Reviews Genetics, 20, 389–403 (2019)
View publicationŽ. Avsec, R. Kreuzhuber, J. Israeli, N. Xu, J. Cheng, A. Shrikumar, A. Banerjee, D. S Kim, T. Beier, L. Urban, A. Kundaje, O. Stegle, J. Gagneur, The Kipoi repository accelerates community exchange and reuse of predictive models for genomics, Nature biotechnology, 2019, PMID: 31138913
View publicationJ. Cheng, M. H. Çelik, Y. D. Nguyen, Ž. Avsec, J. Gagneur, CAGI5 splicing challenge: Improved exon skipping and intron retention predictions with MMSplice, Human Mutation, 2019, PMID: 31070280
View publicationJ. Cheng, T. Y. D. Nguyen, K. J. Cygan, M. H. Celik, W. Fairbrother, Ž. Avsec, J. Gagneur, MMSplice: modular modeling improves the predictions of genetic variant effects on splicing, Genome Biol. 2019, PMID: 30823901
View publication2018
F. Brechtmann, A. Matuseviciute, Ch. Mertes, V. A. Yepez, Z. Avsec, M. Herzog, D. M. Bader, H. Prokisch, J. Gagneur, OUTRIDER: A statistical method for detecting aberrantly expressed genes in RNA sequencing data, AJHG, 2018, PMID: 30503520
View publication2017
Avsec, Z., Barekatain, M., Cheng, J., Gagneur, J. (2017). Modeling positional effects of regulatory sequences with spline transformations increases prediction accuracy of deep neural networks. Bioinformatics doi.10.1093/bioinformatics/btx727
View publicationCheng, J., Maier, K.C., Avsec, Z., Rus, P., and Gagneur, J. (2017). Cis-regulatory elements explain most of the mRNA stability variation across genes in yeast. Rna 23, 1648-1659.
View publicationAit-El-Mkadem, S., Dayem-Quere, M., Gusic, M., Chaussenot, A., Bannwarth, S., François, B., Genin, E.C., Fragaki, K., Volker-Touw, C.L.M., Vasnier, C., et al. Mutations in MDH2, Encoding a Krebs Cycle Enzyme, Cause Early-Onset Severe Encephalopathy. The American Journal of Human Genetics 100, 151-159.
View publication Mattias Backman (until Oct 2019)
Mattias Backman (until Oct 2019)

Mattias Backman (until Oct 2019)
| Class of 2015
Country of origin: Sweden
MSc: Molecular Biology, University of Gothenburg, Sweden
PI: Eckhard Wolf, Gene Center Munich, Dept. of Biochemistry, LMU
2019
Renner S, Martins AS, Streckel E, Braun-Reichhart C, Backman M, Prehn C, Klymiuk N, Bähr A, Blutke A, Landbrecht-Schessl C, Wünsch A, Kessler B, Kurome M, Hinrichs A, Koopmans SJ, Krebs S, Kemter E, Rathkolb B, Nagashima H, Blum H, Ritzmann M, Wanke R, Aigner B, Adamski J, Hrabě de Angelis M, Wolf E; Mild maternal hyperglycemia in INS C93S transgenic pigs causes impaired glucose tolerance and metabolic alterations in neonatal offspring; Dis Model Mech.
View publication2017
Blutke, A., Renner, S., Flenkenthaler, F., Backman, M., Haesner, S., Kemter, E., Landstrom, E., Braun-Reichhart, C., Albl, B., Streckel, E., et al. (2017). The Munich MIDY Pig Biobank – A unique resource for studying organ crosstalk in diabetes. Mol Metab 6, 931-940.
View publication Ivan Bagaric
Ivan Bagaric

Ivan Bagaric
| Class of 2025
Country of origin: Croatia
MSc: Molecular Biosciences, University of Heidelberg, Germany
 Madleen Biggel
Madleen Biggel

Madleen Biggel
| Class of 2026
Country of origin: Germany
BSc: Nutrition Science, TU Munich, Germany
Stingele
 Elena Biselli (Alumna)
Elena Biselli (Alumna)

Elena Biselli (Alumna)
| Class of 2014
Country of origin: Italy
Laurea Magistrale: Physics, University of Rome La Sapienza, Italy
PI: Ulrich Gerland, Faculty of Physics, TU Munich
Project: Characterization and analysis of bacterial behaviour during starvation
I experimentally characterize bacterial behaviour during carbon starvation and quantitatively analyze it using theoretical models. I would like to understand the regulation strategy whereby E.coli cells manage the utilization of a scarce resource and if this strategy is optimized for survival in a fluctuating environment.
2019
S J Schink, E Biselli, C Ammar, U Gerland: Death rate of E. coli during starvation is set by maintenance cost and biomass recycling; Cell Systems, 9, ue 1, 2019, 64-73.e3, ISSN 2405-4712
View publication2018
S J Schink, E Biselli, C Ammar, U Gerland: Maintenance Cost and Biomass Recycling Determine Fitness of E. Coli During Starvation; September 26, 2018, SSRN: https://ssrn.com/abstract=3255561
View publication2017
Biselli, E., Agliari, E., Barra, A., Bertani, F.R., Gerardino, A., De Ninno, A., Mencattini, A., Di Giuseppe, D., Mattei, F., Schiavoni, G., et al. (2017). Organs on chip approach: a tool to evaluate cancer-immune cells interactions. Sci Rep-Uk 7, 12737.
View publication Augusto Borges (Alumnus)
Augusto Borges (Alumnus)

Augusto Borges (Alumnus)
| Class of 2020
Country of origin: Argentinia/Portugal
Argentina
Erwin Frey/ Hernan Lopez-Schier
2023
E. L. Kozak, J. R. Miranda-Rodríguez, A. Borges, K. Dierkes, A. Mineo, F. Pinto-Teixeira, O. Viader-Llargués, J. Solon, O. Chara, H. López-Schier; Quantitative videomicroscopy reveals latent control of cell-pair rotations in vivo. Development 1 May 2023; 150 (9): dev200975.
View publication2022
A S Ceccarelli, A Borges and O Chara: Size matters: tissue size as a marker for a transition between reaction–diffusion regimes in spatio-temporal distribution of morphogens; R. Soc. Open Sci. 9: 211112., 2022
View publication Emily Brieger (Alumna)
Emily Brieger (Alumna)

Emily Brieger (Alumna)
| Class of 2021
Country of origin: Germany
MSc: Physics, Universität Heidelberg
PI: Joachim Rädler, Soft Matter Group, Dept. of Physics, LMU
 Andrew Nelson Butterfield (until Sep 2019)
Andrew Nelson Butterfield (until Sep 2019)
Andrew Nelson Butterfield (until Sep 2019)
| Class of 2019
Country of origin: Germany / UK
MSc: Biology, LMU, Germany
Petra Schwille, MPI of Biochemistry
 Maren Büttner (Alumna)
Maren Büttner (Alumna)

Maren Büttner (Alumna)
| Class of 2014
Country of origin: Germany
MSc: Mathematics, TU Munich, Germany
PI: Fabian Theis, Helmholtz Center Munich, TU Munich
Project: Accounting for noise in single-cell omics
In my PhD, I focus on the analysis of single-cell data and the integration of different data types from several experimental sources. In particular, I focus on data with a large number of features (transcriptomics, esp. RNA-seq) or a large number of samples as in FACS/cyTOF experiments. Mainly the project addresses the need of statistical methods to deal with measurement noise, confounding factors and issues arising from high-dimensionality of the data. Besides, integrating data from different sources demands a robust handling of data processing and possible batch effects.
2019
M. Büttner, Z. Miao, F. A. Wolf, S. A. Teichmann, and F. J. Theis, A test metric for assessing single-cell RNA-seq batch correction. Nature Methods, 16, 43-49 (2019)
View publicationS. Tritschler, M. Büttner, D. S. Fischer, M. Lange, V. Bergen, H. Lickert, F. J. Theis, Concepts and limitations for learning developmental trajectories from single cell genomics, Development 2019 146: dev170506
View publication2017
Buttner, M., Miao, Z., Wolf, A., Teichmann, S.A., and Theis, F.J. (2017). Assessment of batch-correction methods for scRNA-seq data with a new test metric. bioRxiv, 200345.
View publication2016
Haghverdi, L., Büttner, M., Wolf, F.A., Buettner, F., and Theis, F.J. (2016). Diffusion pseudotime robustly reconstructs lineage branching. Nature methods 13, 845-848.
View publication2015
Angerer, P., Haghverdi, L., Büttner, M., Theis, F.J., Marr, C., and Buettner, F. (2015). destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics.
View publication Xueqi Cao (Alumna)
Xueqi Cao (Alumna)

Xueqi Cao (Alumna)
| Class of 2019
Country of origin: China
MSc: Molecular Medicine, University of Jena, Germany
Julien Gagneur, Informatics, TUM
 Purva Choudhary
Purva Choudhary

Purva Choudhary
| Class of 2024
Country of origin: India
MSc: Molecular and Cellular Biology, LMU Munich
 Maíra de Oliveira Torres
Maíra de Oliveira Torres

Maíra de Oliveira Torres
| Class of 2022
Country of origin: Brazil
MSc: Neurology, Federal University of the State of Rio de Janeiro
PI: Johanna Klughammer, Gene Center, LMU
CoPI:
 Timo Denk
Timo Denk
Timo Denk
| Class of 2020
Country of origin: Germany
MSc: Biochemistry, LMUMunich, Germany
Roland Beckmann
Project: Coordination of translation with quality control and mRNA decay in humans
2022
M Narita, T Denk, Y Matsuo, T Sugiyama, C Kikuguchi, S Ito, N Sato, T Suzuki, S Hashimoto, I Machová, P Tesina, R Beckmann & T Inada; A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation, Nat Commun, 13, 6411 (2022)
View publication2020
Thoms, M., Buschauer, R., Ameismeier, M., Koepke, L., Denk, T., Hirschenberger, M., Kratzat, H., Hayn, M., Mackens-Kiani, T., Cheng, J., et al. (2020); Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369, 1249-1255.
View publicationN K Sinha, A Ordureau, K Best, J A Saba, B Zinshteyn, E Sundaramoorthy, A Fulzele, D M Garshott, T Denk, M Thoms, J A Paulo, J W Harper, E J Bennett, R Beckmann, R Green; EDF1 coordinates cellular responses to ribosome collisions; eLife 2020;9:e58828
View publication Alice Fabienne Descoeudres (Alumna)
Alice Fabienne Descoeudres (Alumna)

Alice Fabienne Descoeudres (Alumna)
| Class of 2022
Country of origin: Switzerland
PI: Stefan Canzar, Gene Center, LMU
 Vladana Djakovic
Vladana Djakovic

Vladana Djakovic
| Class of 2024
Country of origin: Bosnia and Herzegovina
MSc: ESG Data Science, LMU Munich
 Maximilian Donsbach
Maximilian Donsbach

Maximilian Donsbach
| Class of 2023
Country of origin: Germany
MSc: Biochemistry, LMU Munich
Project: DNA-Protein-Crosslink Repair, Genome stability
2023
D Yaneva, J L Sparks, M Donsbach, S Zhao, P Weickert, R Bezalel-Buch, J Stingele, J C Walter: The FANCJ helicase unfolds DNA-protein crosslinks to promote their repair, Mol Cell, 2023 Jan 5;83(1):43-56.e10.
View publication Antonia Eicher
Antonia Eicher

Antonia Eicher
| Class of 2023
Country of origin: Germany
MSc: Biochemistry, LMU Munich
 Hanna Esser
Hanna Esser
Hanna Esser
| Class of 2023
Country of origin: UK
MSc: Biochemistry, LMU Munich
 Conrad Fallon
Conrad Fallon

Conrad Fallon
| Class of 2022
Country of origin: UK
MSc: Developmental Biology, University of Manchester
PI: Nicolas Gompel, Dept. of Cell and Developmental Biology, LMU
CoPI:
Project: Modulating a single TFBS in an enhancer: from atomic contacts to variation in enhancer activity
 Bela Frohn
Bela Frohn

Bela Frohn
| Class of 2021
Country of origin: Germany
M.Sc.: Bioinformatics, University of Edinburgh, UK
PI: Petra Schwille, Max Planck Institute of Biochemistry, Martinsried
Project: Bottom-up synthetic biology tries to build minimal living systems from scratch by combining biomolecules such as proteins, lipids and DNA to both understand and engineer life. Up to now, researchers in bottom-up synthetic biology typically have been trying to reconstitute and reduce existing multi-protein systems in vitro by using a minimal number of their components. However, utilising existing proteins is not only inherently contradictory to the idea of bottom-up design as they are themselves not designed from scratch, but also faces severe practical problems as natural proteins are evolutionary optimised for specific in vivo conditions. In my research, I combine computational and experimental tools to on one hand engineer existing protein machineries to fit the specific needs of in vitro reconstitution, and to on the other hand de novo design protein machineries that are potentially easier to understand and control than naturally occurring ones.
2022
P Schwille & B Frohn (2022), Hidden protein functions and what they may teach us; Trends Cell Biol. 32(2):102-109.
View publicationB. Frohn, T. Härtel, J. Cox, P. Schwille (2022): Tracing back variations in archaeal ESCRT-based cell division to protein domain architectures. PLoS One 17(3).
View publication Dandan Guan
Dandan Guan

Dandan Guan
| Class of 2024
Country of origin: China
MSc: Peking Union Medical College, Tsinghua University, China
 Nastaran Hadi Zadeh
Nastaran Hadi Zadeh

Nastaran Hadi Zadeh
| Class of 2025
Country of origin: Iran
years of BSc and 2 years of MSc,, Tehran University of Medical Sciences, Iran
 Jakob Hartmann
Jakob Hartmann

Jakob Hartmann
| Class of 2025
Country of origin: Germany
MSc: Chemistry, LMU Munich, Germany
 Veronika Hepperle
Veronika Hepperle

Veronika Hepperle
| Class of 2026
Country of origin: Germany
BSc: Molecular and Technical Medicine, Furtwangen University of Applied Sciences, Germany
 Hendrik Jock
Hendrik Jock
Hendrik Jock
| Class of 2024
Country of origin: Germany
MSc: Biochemistry, LMU Munich
 Florian Katzmeier (Alumnus)
Florian Katzmeier (Alumnus)

Florian Katzmeier (Alumnus)
| Class of 2019
Country of origin: Germany
MSc: Physics, Technische Universität München, Germany
Fritz Simmel, Physics, TUM
 Alena Khmelinskaia (Alumna)
Alena Khmelinskaia (Alumna)

Alena Khmelinskaia (Alumna)
| Class of 2014
Country of origin: Portugal
MSc: Biochemistry, University of Lisbon, Portugal
PI: Petra Schwille, Max Planck Institute of Biochemistry, Martinsried
CoPI: Joachim Rädler, Faculty of Physics, LMU Munich
Project: Development of synthetic DNA-based membrane scaffolds
In this project DNA-origami based synthetic nanostructures are being produced to mimic the mechanism of action of scaffolding proteins. Here, we aim to unravel the minimal requirements for lipid membrane binding, self-organization in 2D as well as recognition or induction of membrane curvature.
2019
H. Eto, N. Soga, H. G. Franquelim, P. Glock, A. Khmelinskaia, L. Kai, M. Heymann, H. Noji and P. Schwille; Design of Sealable Custom-Shaped Cell Mimicries Based on Self- Assembled Monolayers on CYTOP Polymer, ACS Appl. Mater. Interfaces 2019, 11, 21372−21380
View publicationS. Kempter, A. Khmelinskaia, M. T. Strauss, P. Schwille, R. Jungmann, T. Liedl, W. Bae; Single Particle Tracking and Super-Resolution Imaging of Membrane-Assisted Stop-and-Go Diffusion and Lattice Assembly of DNA Origami; ACS Nano, 2019, 132, 996-1002
View publication2018
Franquelim, H.G., Khmelinskaia, A., Sobczak, J.P., Dietz, H., and Schwille, P. (2018). Membrane sculpting by curved DNA origami scaffolds. Nat Commun 9, 811.
View publicationA. Khmelinskaia, J. Mücksch, E. P. Petrov, H. G. Franquelim. P. Schwille; Control of Membrane Binding and Diffusion of Cholesteryl-Modified DNA Origami Nanostructures by DNA Spacers; Langmuir, 2018, 34, 49, 14921-14931
View publicationA. Khmelinskaia, J. Mücksch, F. Conci, G. Chwastek, P. Schwille; FCS Analysis of Protein Mobility on Lipid Monolayers, Open Archive Published: March 28, 2018 DOI:https://doi.org/10.1016/j.bpj.2018.02.031
View publicationW. Ye, S. Celiksoy, A. Jakab, A. Khmelinskaia, T. Heermann, A. Raso, S. V. Wegner, G. Rivas, P. Schwille, R. Ahijado-Guzmán, C. Sönnichsen; Plasmonic Nanosensors Reveal a Height Dependence of MinDE Protein Oscillations on Membrane Features, J. Am. Chem. Soc.2018, 140, 51
View publication2017
Vogel, S.K., Greiss, F., Khmelinskaia, A., and Schwille, P. (2017). Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife 6.
View publication2016
Khmelinskaia, A., Franquelim, H.G., Petrov, E.P., and Schwille, P. (2016). Effect of anchor positioning on binding and diffusion of elongated 3D DNA nanostructures on lipid membranes. J Phys D Appl Phys 49.
View publication Bernhard Kirchmair
Bernhard Kirchmair

Bernhard Kirchmair
| Class of 2026
Country of origin: Germany
BSc: Physics, LMU Munich, Germany
 Lara Charlotte Kopp
Lara Charlotte Kopp

Lara Charlotte Kopp
| Class of 2025
Country of origin: Germany
MSc: Biomolecular Engineering, TU Darmstadt, Germany
 Ching Yee Leung
Ching Yee Leung

Ching Yee Leung
| Class of 2019
Country of origin: Hong Kong
MSc: Physics, Chinese University of Hong Kong, China
Erwin Frey, Physics, LMU
Project: Pattern formation
Pattern formation is crucial in biology as it offers a theoretical framework for understanding complex and spatially organized patterns in biological systems. Reaction-diffusion models are one of the most successful theories in this regard, with the 2-component mass conserving reaction diffusion model (2C-McRD) being the simplest. The local equilibria theory can well describe 2C-McRD, but more complex systems with additional components and conserved species have a significant gap between the 2C-McRD and multiple-component McRD (MC-McRD). Simple models like the Min system have been modified, but it leads to an increase in complexity and requires numerical simulations. To address this gap, the goal is to generalize the local equilibria theory for more complex systems by identifying important subspaces in the phase space of MC-McRD systems and classifying their behavior, which is different from the standard approach of reducing model complexity while retaining the main features.
2020
M. C. Wigbers, F. Brauns, C. Y. Leung and E. Frey; Flow Induced Symmetry Breaking in a Conceptual Polarity Model, Cells 2020, 9(6), 1524;
View publication Garp Linder
Garp Linder

Garp Linder
| Class of 2023
Country of origin: Germany
MSc: Biology, LMU Munich
Project: DNA shape read-out of the INO80 chromatin remodeler revealed by single-molecule imaging
2021
O’Neill, L., Linder, G., van Buuren, M., & von Bayern, A.M.P. (2021). New Caledonian crows and hidden causal agents revisited; Animal Behavior and Cognition, 8(2), 166-189.
View publication Mohammad Mokhtari
Mohammad Mokhtari

Mohammad Mokhtari
| Class of 2022
Country of origin: Iran
PI: Johanna Klughammer, Gene Center, LMU
CoPI:
Project: Spatio-molecular characterization of hepatocellular carcinoma immunotherapy in a mouse model
 Mariam Museridze (Alumna)
Mariam Museridze (Alumna)
Mariam Museridze (Alumna)
| Class of 2020
Country of origin: Georgia
MSc: Biology, LMU Munich, Germany
Nicolas Gompel
2020
Xin, Y., Poul, Y. Le, Ling, L., Museridze, M., Mühling, B., Jaenichen, R., Osipova, E., and Gompel, N.: Enhancer evolutionary co-option through shared chromatin accessibility input, Proc. Natl. Acad. Sci. 117, 20636–20644 (2020).
View publicationPoul, Y. Le, Xin, Y., Ling, L., Mühling, B., Jaenichen, R., Hörl, D., Bunk, D., Harz, H., Leonhardt, H., Wang, Y., et al., & Museridze M, Gompel, N: Regulatory encoding of quantitative variation in spatial activity of a Drosophila enhancer. Sci. Adv. 6, eabe2955 (2020).
View publication Akshay Patel (external)
Akshay Patel (external)

Akshay Patel (external)
| Class of 2021
Country of origin: USA
PI: Stefan Canzar, Gene Center, Dept. of Biochemistry, LMU
 Chandhana Prakash
Chandhana Prakash

Chandhana Prakash
| Class of 2025
Country of origin: India
MSc: Biological Sciences, Indian Institute of Science Education and Research Thiruvananthapuram (IISER Thiruvananthapuram), India
 Michael Pries
Michael Pries
Michael Pries
| Class of 2023
Country of origin: Germany
MSc: Molecular and Cellular Biology, LMU Munich
 Elias Reutelsterz
Elias Reutelsterz

Elias Reutelsterz
| Class of 2026
Country of origin: Germany
BSc: Business Mathematics, University of Mannheim, Germany
 Andreas Riedlberger
Andreas Riedlberger

Andreas Riedlberger
| Class of 2025
Country of origin: Germany
MSc: Biology, University of Freiburg
 Cristian Rosales (Alumnus)
Cristian Rosales (Alumnus)

Cristian Rosales (Alumnus)
| Class of 2021
Country of origin: Columbia
PI: Roland Beckmann, Gene Center, Dept. of Biochemistriy, LMU
 Dina Sophia Schnapka
Dina Sophia Schnapka

Dina Sophia Schnapka
| Class of 2025
Country of origin: Germany
MSc: Chemistry, LMU Munich, Germany
 Sophia Schwojer
Sophia Schwojer

Sophia Schwojer
| Class of 2024
Country of origin: Germany
MSc: Biochemistry, LMU Munich
2022
Stafford CA, Gassauer AM, de Oliveira Mann CC, Tanzer MC, Fessler E, Wefers B, Nagl D, Kuut G, Sulek K, Vasilopoulou C, Schwojer SJ, Wiest A, Pfautsch MK, Wurst W, Yabal M, Fröhlich T, Mann M, Gisch N, Jae LT, Hornung V.
Nature. 2022 Aug 24. doi: 10.1038/s41586-022-05125-x.
 Bachuki Shashikadze (Alumnus)
Bachuki Shashikadze (Alumnus)

Bachuki Shashikadze (Alumnus)
| Class of 2021
Country of origin: Georgia
MSc Chemistry, Université Paris-Saclay, France
2022
Stirm, M., L. Fonteyne, B. Shashikadze, J. Stöckl, M. Kurome, B. Keßler, V. Zakhartchenko, E. Kemter, H. Blum, G. J. Arnold, K. Matiasek, R. Wanke, W. Wurst, H. Nagashima, F. Knieling, M. Walter, C. Kupatt, T. Fröhlich, N. Klymiuk and E. Wolf (2022). „Pig models for Duchenne muscular dystrophy – from disease mechanisms to validation of new diagnostic and therapeutic concepts.“ Neuromuscular Disorders. DOI:https://doi.org/10.1016/j.nmd.2022.04.005
View publication2021
Flenkenthaler, F., E. Ländström, B. Shashikadze, M. Backman, A. Blutke, J. Philippou-Massier, S. Renner, M. Hrabe de Angelis, R. Wanke, H. Blum, G. J. Arnold, E. Wolf and T. Fröhlich (2021): Differential Effects of Insulin-Deficient Diabetes Mellitus on Visceral vs. Subcutaneous Adipose Tissue-Multi-omics Insights From the Munich MIDY Pig Model, Frontiers in medicine 8: 751277-751277.
View publicationShashikadze, B., F. Flenkenthaler, J. Stöckl, L. Valla, S. Renner, E. Kemter, E. Wolf and T. Fröhlich (2021): „Developmental Effects of (Pre-)Gestational Diabetes on Offspring: Systematic Screening Using Omics Approaches.“ Genes 12: 1991.
View publicationStirm, M., L. M. Fonteyne, B. Shashikadze, M. Lindner, M. Chirivi, A. Lange, C. Kaufhold, C. Mayer, I. Medugorac, B. Kessler, M. Kurome, V. Zakhartchenko, A. Hinrichs, E. Kemter, S. Krause, R. Wanke, G. J. Arnold, G. Wess, H. Nagashima, M. Hrabĕ de Angelis, F. Flenkenthaler, L. A. Kobelke, C. Bearzi, R. Rizzi, A. Bähr, S. Reese, K. Matiasek, M. C. Walter, C. Kupatt, S. Ziegler, P. Bartenstein, T. Fröhlich, N. Klymiuk, A. Blutke and E. Wolf (2021). „A scalable, clinically severe pig model for Duchenne muscular dystrophy.“ Disease Models & Mechanisms 14(12).
View publication Niu Shuangshuang
Niu Shuangshuang

Niu Shuangshuang
| Class of 2019
Country of origin: China
MSc: Biochemistry and Molecular Biology, University of Chinese Academy of Sciences, China
Roland Beckmann, Biochemistry, LMU
 Robert Strasser
Robert Strasser

Robert Strasser
| Class of 2022
Country of origin: Germany
MSc: Biology, LMU Munich
PI: Johanna Klughammer, Gene Center, LMU
CoPI:
Project: Chemotherapeutic impact on the immune microenvironment in metastatic breast cancer:
In women, breast cancer is the most common type of cancer, having a global incidence of over two million in 2020. In a fifth of all breast cancer diagnoses, liver metastases occur, which, despite treatment with hormone- and/or chemotherapy, lead to a 5-year survival rate of only 27%. In collaboration with researcher at the Broad Institute we investigate the heterogenous cellular ecosystem of liver metastasis at the level of individual cells, using single-nucleus RNA sequencing (snRNA-seq), and additionally on a spatial level, using Multiplexed error-robust fluorescence in situ hybridization (MERFISH). We assess and compare molecular and compositional differences between metastasis of patients that have or have not been treated with chemotherapy to better understand how chemotherapy might impact the effectiveness of immunotherapy as a complementary line of therapy
 Yee Man Tam
Yee Man Tam
Yee Man Tam
| Class of 2026
Country of origin: China
BSc: Biochemistry, The University of Hong Kong, China
 Taja Vatovec
Taja Vatovec

Taja Vatovec
| Class of 2025
Country of origin: Slovenia
MSc: Molecular Biosciences, University of Heidelberg, Germany
 Tatsiana Volkava
Tatsiana Volkava

Tatsiana Volkava
| Class of 2026
Country of origin: Belarus
BSc: Applied Mathematics and Physics, Moscow Institute of Physics and Technology, Russia
 Victoria Vyboishchikov Oslonovitch
Victoria Vyboishchikov Oslonovitch

Victoria Vyboishchikov Oslonovitch
| Class of 2026
Country of origin: Spain
BSc: Biology, University of Tübingen, Germany
Stingele
 Jan Watter
Jan Watter

Jan Watter
| Class of 2022
Country of origin: Germany
M.Sc.: Computational Science and Engineering, TUM
PI: Johanna Klughammer, Gene Center, LMU
CoPI:
Project: Methods development for high-dimensional spatio-molecular data analysis
Single cell, spatial and omics methods provide high-dimensional molecular measurements of cellular ecosystems at unprecedented scale and resolution. A key challenge is the meaningful interpretation of these information-rich biological data, with the goal to improve our understanding of cellular communication, interaction, and collective behaviour on a molecular level.
This project builds on our python-based framework called TACCO (Transfer of Annotations to Cells and their COmbinations) to improve and extend its functionalities. Specifically, it tackles a particularly challenging problem which is to transfer information from the ever-increasing wealth of scRNAseq data to spatial proteomics data such as CODEX, overcoming technical as well as biological discrepancies between these different modalities.
 Melanie Weiss
Melanie Weiss

Melanie Weiss
| Class of 2024
Country of origin: Germany
MSc: Medical Biology, University of Duisburg Essen
 Jan Willeke
Jan Willeke

Jan Willeke
| Class of 2024
Country of origin: Germany
M.Sc. Physics, LMU Munich
 Gang (Jack) Yu
Gang (Jack) Yu

Gang (Jack) Yu
| Class of 2024
Country of origin: China
MS: Johns Hopkins University (Baltimore, Maryland, USA)
Project: Prediction of the translational effects of Single Nucleotide Polymorphisms (SNPs) using machine learning
2022
Nakamoto, M., Tao, J., & Yu, J. Ageas: Automated machine learning based genetic regulatory element extraction system. bioRxiv.2022
View publication Patrick Zechner
Patrick Zechner

Patrick Zechner
| Class of 2026
Country of origin: Germany
BSc: Biology, LMU München, Germany
 Daniel Zhou
Daniel Zhou

Daniel Zhou
| Class of 2026
BSc: Physics, TU Munich, Germany
Frey
 Alexander Ziepke (Alumnus)
Alexander Ziepke (Alumnus)

Alexander Ziepke (Alumnus)
| Class of 2019
Country of origin: Germany
MSc: Physics, Technische Universität Berlin, Germany
Erwin Frey, Physics, LMU

 Agathe Jouneau
Agathe Jouneau
 Shuhan Yang
Shuhan Yang